Welcome to Soybean LPA Gene Network
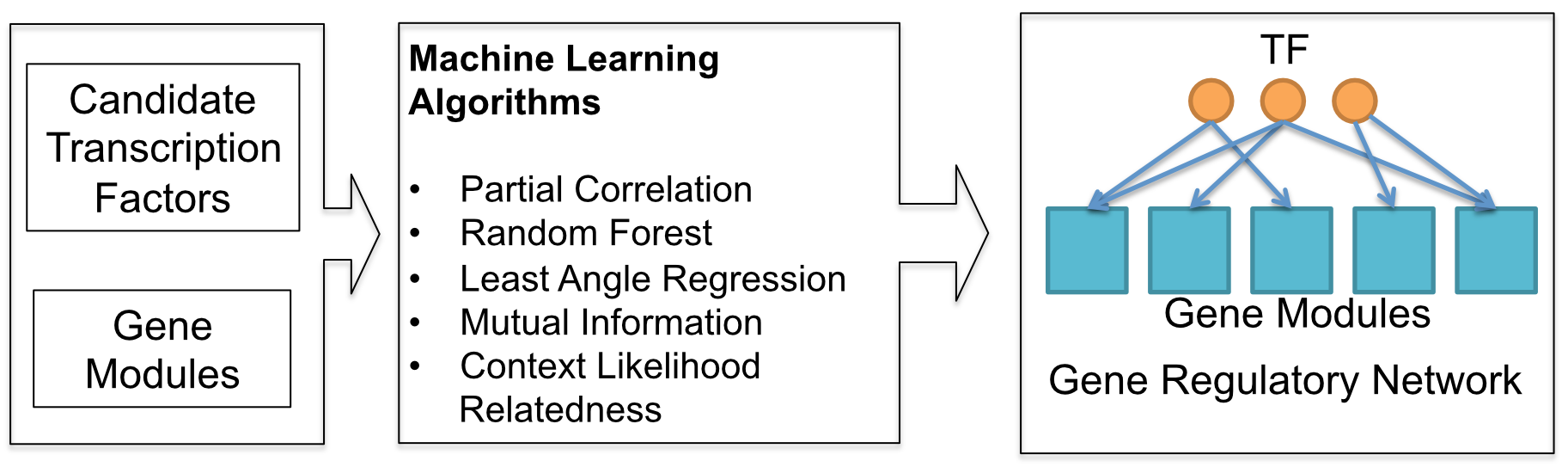
We applied five machine learning methods to learn gene regulatory networks from RNAseq data generated during soybean seed development.
These machine learning methods include:
- Mutual Information (ARACNE)
- Random Forest (RF)
- Context Likelihood Relatedness (CLR)
- Least Angle Regression (LARS)
- Partial Correlation (PCOR)
Usage
Input data
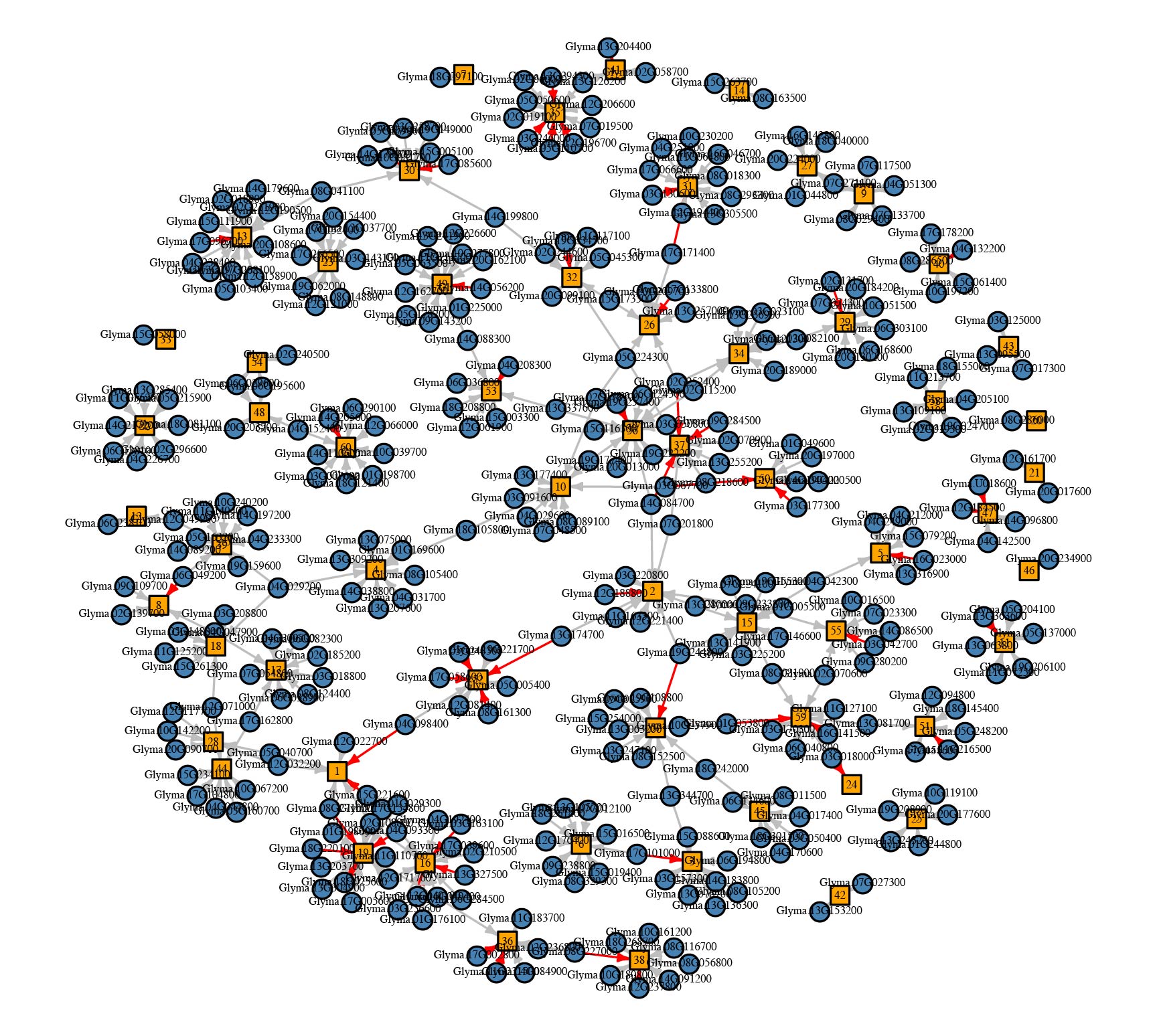
Input data include gene expression from a time series experiment or multiple tissue types. Data should be in matrix format. With each row represents the expression pattern of a transcription factor (TF), or a gene module. Gene module can be obtained using clustering method. A sapmle input data is provided for 30 transcription factors and 60 gene modules. Each row is the expression data for one gene or a module. Column 1 is the gene names and module names. Other columns are experimental conditions.
Perform network analysis
Name the input data as TFandModule.csv, run the R code using Rstudio or Rscript command. For example, if you want to run the ARACNE method, use the following command.
Rscript NetworkLearning_ARACNE.R
This will create a file called “aracne_network_edges.csv” in the current working directory.
Output
Output visualization is performed using R package igraph