Regulatory Genomics
We are interested in understanding gene regulation using network biology and machine learning approaches. We have developed several computational tools to analyze and predict key regulators of gene expression by integrating gene expression, protein-DNA interaction and epigenomic regulations.
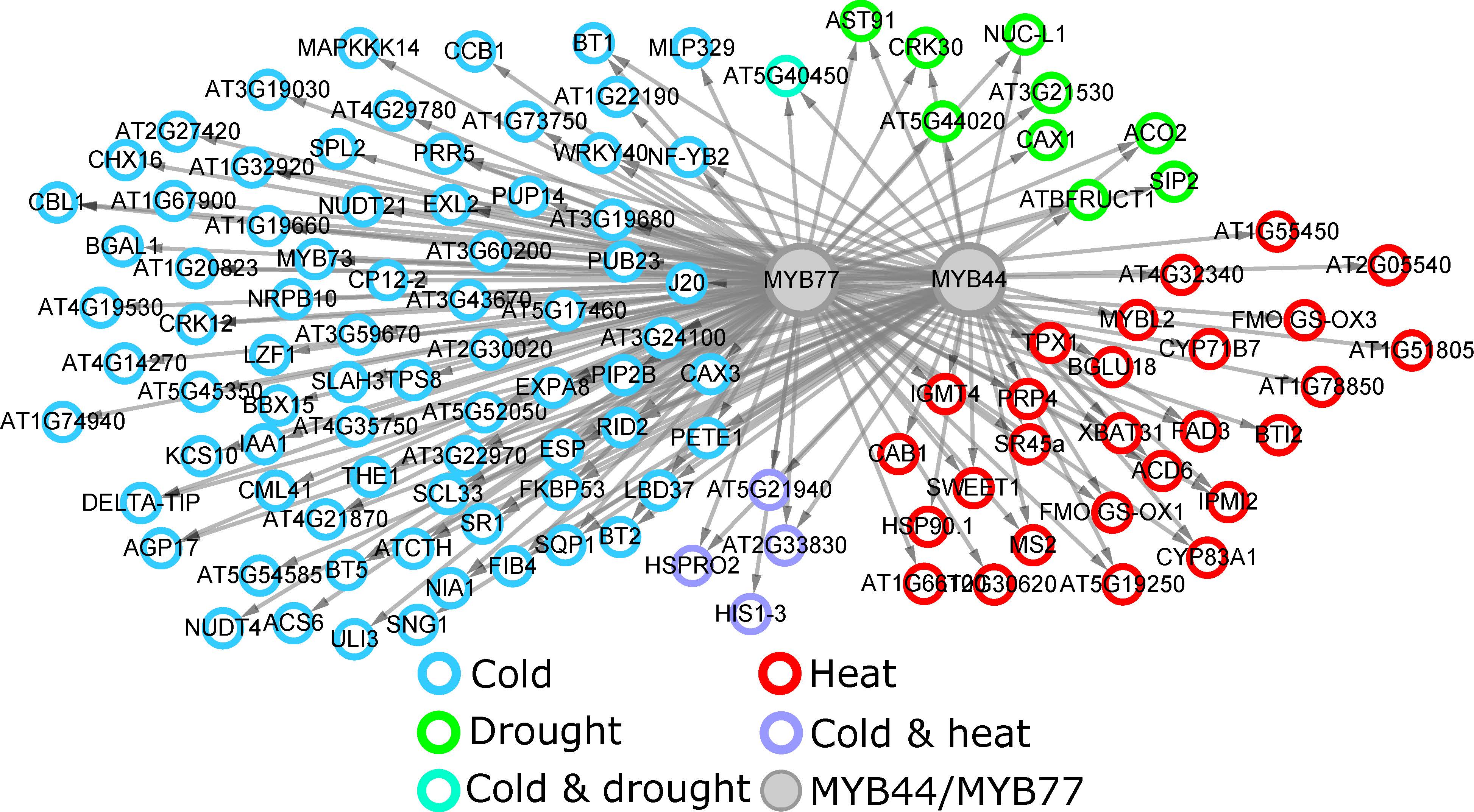
ConSReg is a machine learning pipeline which select best candidate regulatory gene by integrating DAP-seq, RNA-seq and ATAC-seq data in Arabidopsis. ConSReg have been tested using both bulk RNA-seq and single cell RNA-seq data. Based on benchmark using known regulatory transcription factors, ConSReg performs better than commonly used enrichment test and provide higher ranking for known TFs. We also found that including ATAC-seq in the model substantially improves the model performance. [publication] and [software]
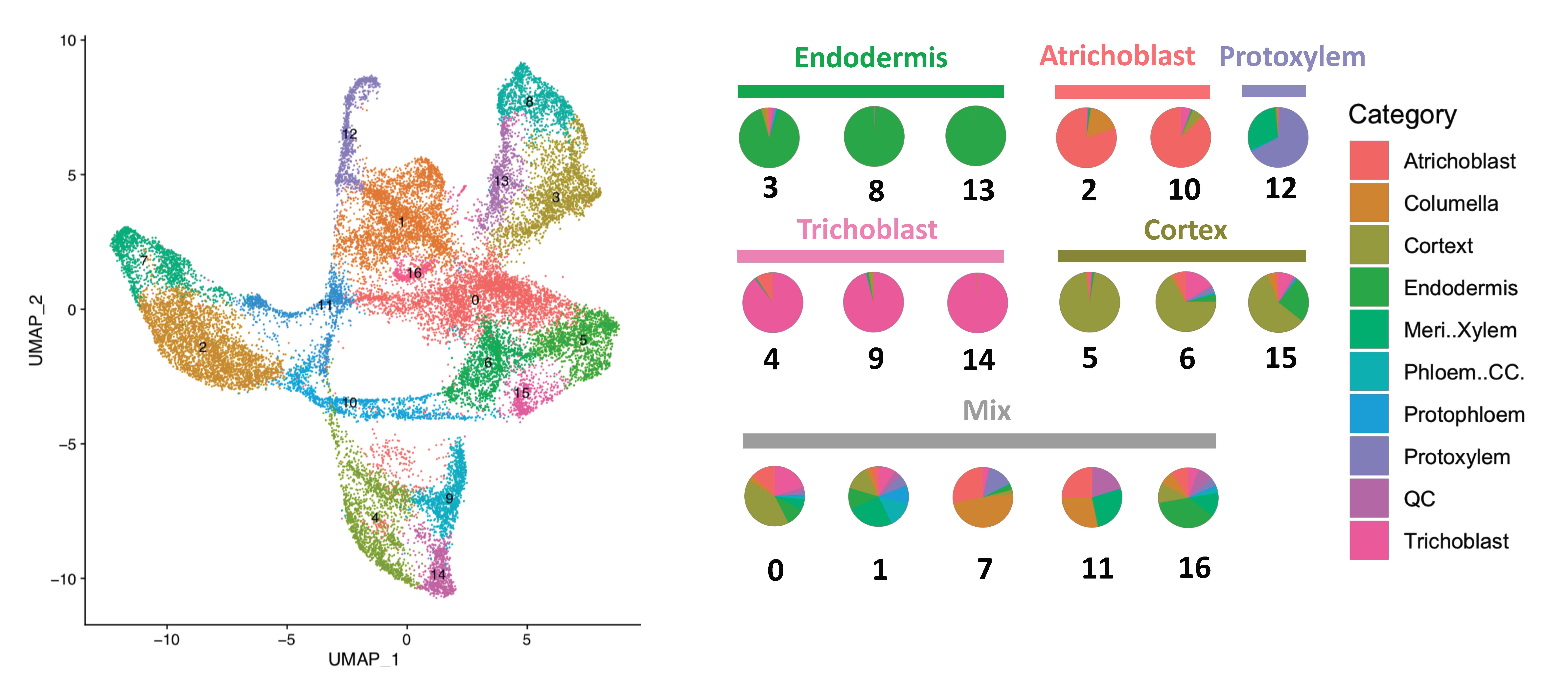
Single Cell SPMarker is a collection of machine-learning methods that can be used to select cell type marker genes in single cell RNA-seq data. We tested multiple feature selection approaches to determine candidate genes that can be used to assign cell types in single cell sequencing data in plant roots. We found that a random forest method + SHAP based feature selection performs best among all approaches tested. We also provide lists of new marker genes that have not been identified before using bulk RNA-seq or microarray experiments in Arabidopsis roots. [biorxiv], [link to code]
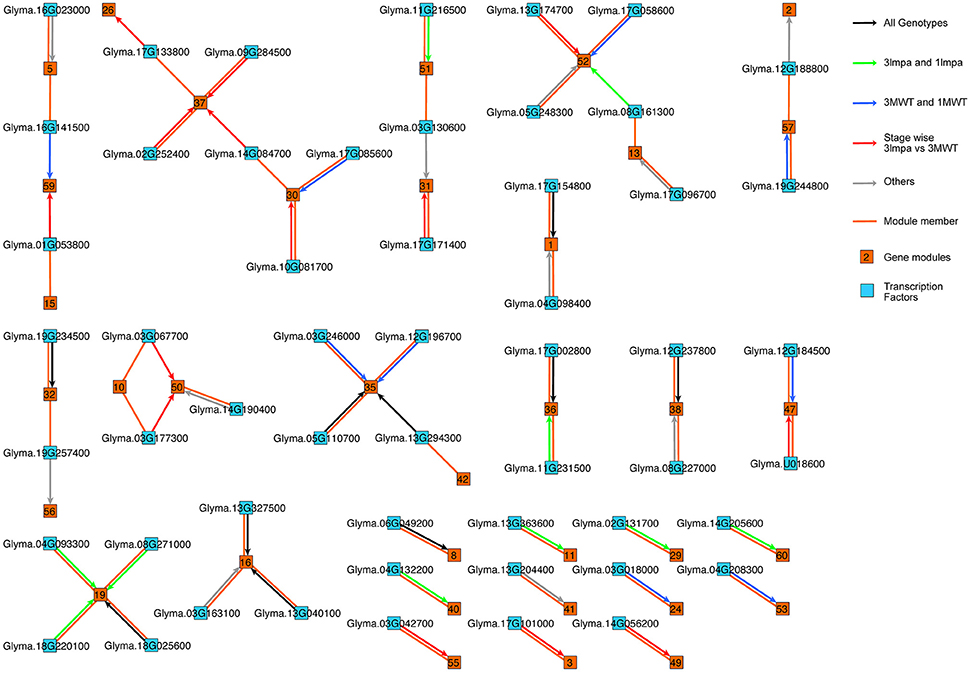
Comparing time series gene expression data across plant species. In this work, we developed a network module finding method to identify conserved co-expression modules between different plant species without matching the tissue types between different species. Figure 3. [publication] [link to code]
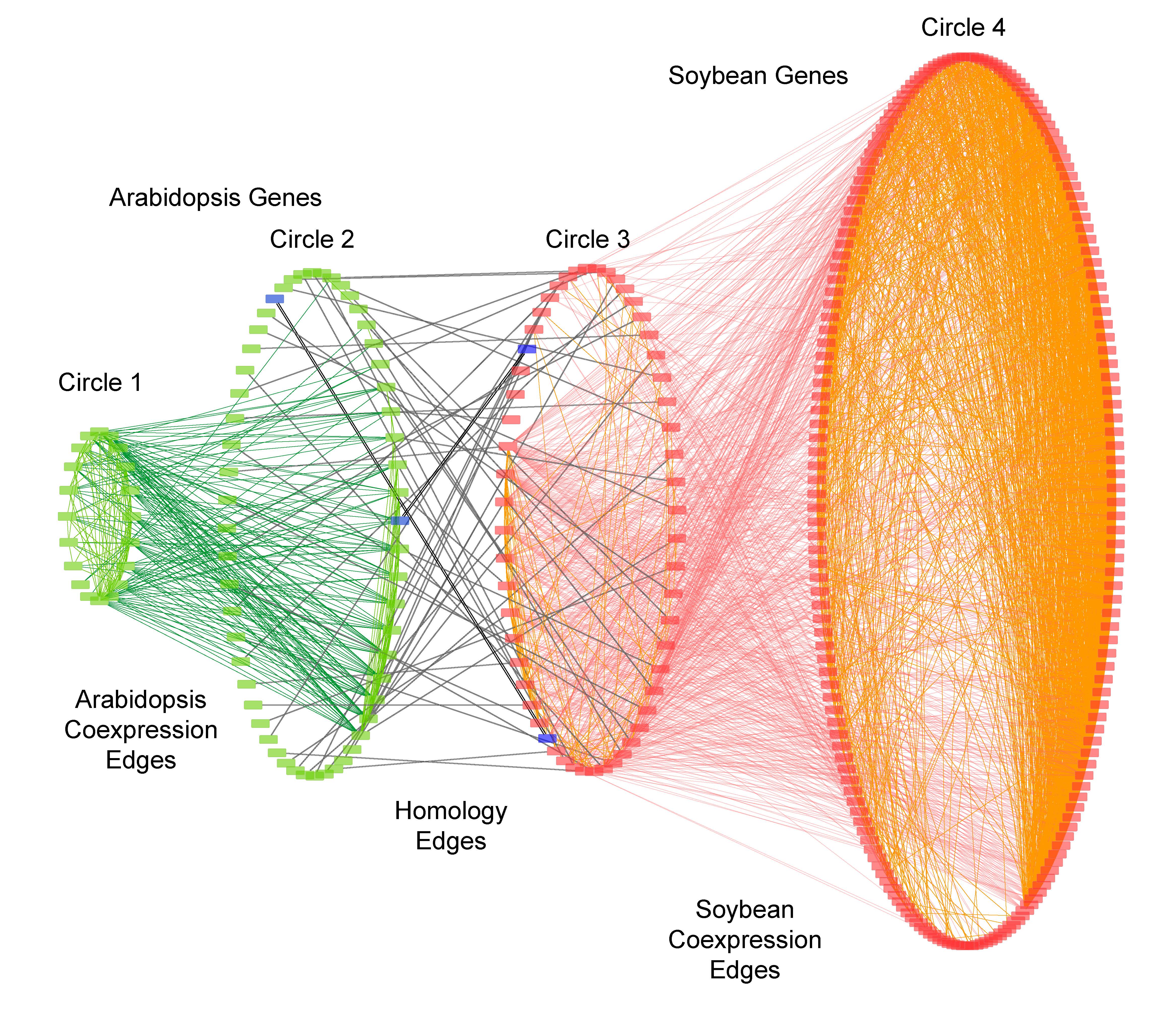
Ensemble network prediction using gene expression data. We have developed supervised and unsupervised machine learning methods to infer regulatory networks from gene expression data for both Arabidopsis and soybeans. We are interested in applying these approaches to gene expression data in other plant species. [publication]